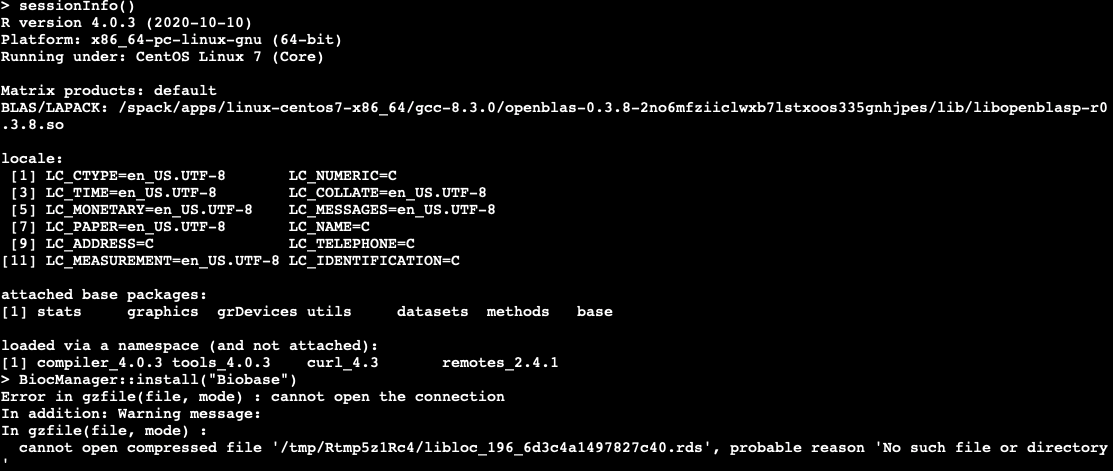
Hi, could anyone please help me with the r error when I want to install any packages, I am taking Biobase as an example below
Skipping 1 packages not available: Biobase
Installing 26 packages: RcppArmadillo, matrixStats, Rcpp, conquer, MatrixModels, SparseM, utf8, crayon, vctrs, rlang, pillar, lifecycle, fansi, ellipsis, colorspace, viridisLite, R6, farver, quantreg, mcmc, tibble, isoband, digest, MCMCpack, ggplot2, nnls
Installing packages into ‘/project/rohs_102/jiaweih/projects/R/packages’
(as ‘lib’ is unspecified)
Error: Failed to install 'unknown package' from GitHub:
unable to create temporary directory ‘/tmp/Rtmp5z1Rc4/downloaded_packages’
In addition: Warning messages:
1: In dir.create(tmpd) :
cannot create dir '/tmp/Rtmp5z1Rc4/downloaded_packages', reason 'No such file or directory'
2: In file(con, "wb") :
cannot open file '/tmp/Rtmp5z1Rc4/file5f4c439189e6': No such file or directory
can anyone help me? Thanks in advance