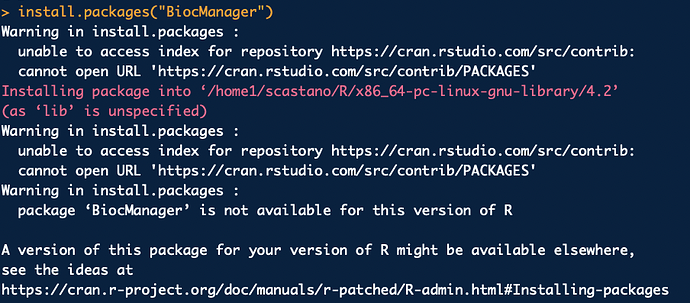
Hello,
I have not been able to install the BiocManager onto RStudio. An error message appears stating that the package is not available with the current version of R. I have tried opening an older version but to no avail.
Any help is appreciated.
Sofi
@scastano I made a change to the RStudio app that should resolve the Bioconductor connection issue. Please try installing a Bioconductor package from within RStudio again.
If needed, another workaround is to set this option:
options(BioC_mirror = "http://bioconductor.org", repos = "http://cran.r-project.org")
Hey Derek,
Thanks for getting back to me. I tried to install again and was not able to. Below I will post the error message. I tried to follow the instructions to where it says to make sure to download openssl, but I cannot seem to install and load that one either. Sorry this is such a hassle.
- installing source package ‘png’ …
** package ‘png’ successfully unpacked and MD5 sums checked
** using staged installation
** libs
/spack/apps/gcc/11.3.0/bin/gcc -I"/spack/2206/apps/linux-centos7-x86_64_v3/gcc-11.3.0/r-4.2.1-gof522x/rlib/R/include" -DNDEBUG -I/usr/local/include libpng-config --cflags -fpic -g -O2 -c read.c -o read.o
/spack/apps/gcc/11.3.0/bin/gcc -I"/spack/2206/apps/linux-centos7-x86_64_v3/gcc-11.3.0/r-4.2.1-gof522x/rlib/R/include" -DNDEBUG -I/usr/local/include libpng-config --cflags -fpic -g -O2 -c write.c -o write.o
/spack/apps/gcc/11.3.0/bin/gcc -shared -L/spack/2206/apps/linux-centos7-x86_64_v3/gcc-11.3.0/r-4.2.1-gof522x/rlib/R/lib -L/spack/2206/apps/linux-centos7-x86_64_v3/gcc-11.3.0/r-4.2.1-gof522x/rlib/R/lib -Wl,-rpath,/spack/2206/apps/linux-centos7-x86_64_v3/gcc-11.3.0/r-4.2.1-gof522x/rlib/R/lib -o png.so read.o write.o -L/spack/2206/apps/linux-centos7-x86_64_v3/gcc-11.3.0/libpng-1.6.37-m3jd5ur/lib -lpng16 -lm -lz -lm -L/spack/2206/apps/linux-centos7-x86_64_v3/gcc-11.3.0/r-4.2.1-gof522x/rlib/R/lib -lR
/bin/ld: cannot find -lz
collect2: error: ld returned 1 exit status
make: *** [/spack/2206/apps/linux-centos7-x86_64_v3/gcc-11.3.0/r-4.2.1-gof522x/rlib/R/share/make/shlib.mk:10: png.so] Error 1
ERROR: compilation failed for package ‘png’
- removing ‘/home1/scastano/R/x86_64-pc-linux-gnu-library/4.2/png’
- installing source package ‘openssl’ …
** package ‘openssl’ successfully unpacked and MD5 sums checked
** using staged installation
Using PKG_CFLAGS=
--------------------------- [ANTICONF] --------------------------------
Configuration failed because openssl was not found. Try installing:
- deb: libssl-dev (Debian, Ubuntu, etc)
- rpm: openssl-devel (Fedora, CentOS, RHEL)
- csw: libssl_dev (Solaris)
- brew: openssl (Mac OSX)
If openssl is already installed, check that ‘pkg-config’ is in your
PATH and PKG_CONFIG_PATH contains a openssl.pc file. If pkg-config
is unavailable you can set INCLUDE_DIR and LIB_DIR manually via:
R CMD INSTALL --configure-vars=‘INCLUDE_DIR=… LIB_DIR=…’
-------------------------- [ERROR MESSAGE] ---------------------------
tools/version.c:1:10: fatal error: openssl/opensslv.h: No such file or directory
1 | #include <openssl/opensslv.h>
| ^~~~~~~~~~~~~~~~~~~~
compilation terminated.
ERROR: configuration failed for package ‘openssl’
- removing ‘/home1/scastano/R/x86_64-pc-linux-gnu-library/4.2/openssl’
- installing source package ‘RCurl’ …
** package ‘RCurl’ successfully unpacked and MD5 sums checked
** using staged installation
checking for curl-config… /spack/2206/apps/linux-centos7-x86_64_v3/gcc-11.3.0/curl-7.83.0-hjubfti/bin/curl-config
checking for gcc… gcc
checking whether the C compiler works… no
configure: error: in /tmp/RtmpU0HU7F/R.INSTALL566479fbcb0/RCurl': configure: error: C compiler cannot create executables See config.log’ for more details
ERROR: configuration failed for package ‘RCurl’
- removing ‘/home1/scastano/R/x86_64-pc-linux-gnu-library/4.2/RCurl’
- installing source package ‘XML’ …
** package ‘XML’ successfully unpacked and MD5 sums checked
** using staged installation
checking for gcc… gcc
checking whether the C compiler works… no
configure: error: in /tmp/Rtmp6kQDvt/R.INSTALL61c19a24337/XML': configure: error: C compiler cannot create executables See config.log’ for more details
ERROR: configuration failed for package ‘XML’
- removing ‘/home1/scastano/R/x86_64-pc-linux-gnu-library/4.2/XML’
- installing source package ‘XVector’ …
** using staged installation
** libs
/spack/apps/gcc/11.3.0/bin/gcc -I"/spack/2206/apps/linux-centos7-x86_64_v3/gcc-11.3.0/r-4.2.1-gof522x/rlib/R/include" -DNDEBUG -I’/home1/scastano/R/x86_64-pc-linux-gnu-library/4.2/S4Vectors/include’ -I’/home1/scastano/R/x86_64-pc-linux-gnu-library/4.2/IRanges/include’ -I/usr/local/include -fpic -g -O2 -c IRanges_stubs.c -o IRanges_stubs.o
/spack/apps/gcc/11.3.0/bin/gcc -I"/spack/2206/apps/linux-centos7-x86_64_v3/gcc-11.3.0/r-4.2.1-gof522x/rlib/R/include" -DNDEBUG -I’/home1/scastano/R/x86_64-pc-linux-gnu-library/4.2/S4Vectors/include’ -I’/home1/scastano/R/x86_64-pc-linux-gnu-library/4.2/IRanges/include’ -I/usr/local/include -fpic -g -O2 -c Ocopy_byteblocks.c -o Ocopy_byteblocks.o
/spack/apps/gcc/11.3.0/bin/gcc -I"/spack/2206/apps/linux-centos7-x86_64_v3/gcc-11.3.0/r-4.2.1-gof522x/rlib/R/include" -DNDEBUG -I’/home1/scastano/R/x86_64-pc-linux-gnu-library/4.2/S4Vectors/include’ -I’/home1/scastano/R/x86_64-pc-linux-gnu-library/4.2/IRanges/include’ -I/usr/local/include -fpic -g -O2 -c RDS_random_access.c -o RDS_random_access.o
/spack/apps/gcc/11.3.0/bin/gcc -I"/spack/2206/apps/linux-centos7-x86_64_v3/gcc-11.3.0/r-4.2.1-gof522x/rlib/R/include" -DNDEBUG -I’/home1/scastano/R/x86_64-pc-linux-gnu-library/4.2/S4Vectors/include’ -I’/home1/scastano/R/x86_64-pc-linux-gnu-library/4.2/IRanges/include’ -I/usr/local/include -fpic -g -O2 -c R_init_XVector.c -o R_init_XVector.o
/spack/apps/gcc/11.3.0/bin/gcc -I"/spack/2206/apps/linux-centos7-x86_64_v3/gcc-11.3.0/r-4.2.1-gof522x/rlib/R/include" -DNDEBUG -I’/home1/scastano/R/x86_64-pc-linux-gnu-library/4.2/S4Vectors/include’ -I’/home1/scastano/R/x86_64-pc-linux-gnu-library/4.2/IRanges/include’ -I/usr/local/include -fpic -g -O2 -c S4Vectors_stubs.c -o S4Vectors_stubs.o
/spack/apps/gcc/11.3.0/bin/gcc -I"/spack/2206/apps/linux-centos7-x86_64_v3/gcc-11.3.0/r-4.2.1-gof522x/rlib/R/include" -DNDEBUG -I’/home1/scastano/R/x86_64-pc-linux-gnu-library/4.2/S4Vectors/include’ -I’/home1/scastano/R/x86_64-pc-linux-gnu-library/4.2/IRanges/include’ -I/usr/local/include -fpic -g -O2 -c SharedDouble_class.c -o SharedDouble_class.o
/spack/apps/gcc/11.3.0/bin/gcc -I"/spack/2206/apps/linux-centos7-x86_64_v3/gcc-11.3.0/r-4.2.1-gof522x/rlib/R/include" -DNDEBUG -I’/home1/scastano/R/x86_64-pc-linux-gnu-library/4.2/S4Vectors/include’ -I’/home1/scastano/R/x86_64-pc-linux-gnu-library/4.2/IRanges/include’ -I/usr/local/include -fpic -g -O2 -c SharedInteger_class.c -o SharedInteger_class.o
/spack/apps/gcc/11.3.0/bin/gcc -I"/spack/2206/apps/linux-centos7-x86_64_v3/gcc-11.3.0/r-4.2.1-gof522x/rlib/R/include" -DNDEBUG -I’/home1/scastano/R/x86_64-pc-linux-gnu-library/4.2/S4Vectors/include’ -I’/home1/scastano/R/x86_64-pc-linux-gnu-library/4.2/IRanges/include’ -I/usr/local/include -fpic -g -O2 -c SharedRaw_class.c -o SharedRaw_class.o
/spack/apps/gcc/11.3.0/bin/gcc -I"/spack/2206/apps/linux-centos7-x86_64_v3/gcc-11.3.0/r-4.2.1-gof522x/rlib/R/include" -DNDEBUG -I’/home1/scastano/R/x86_64-pc-linux-gnu-library/4.2/S4Vectors/include’ -I’/home1/scastano/R/x86_64-pc-linux-gnu-library/4.2/IRanges/include’ -I/usr/local/include -fpic -g -O2 -c SharedVector_class.c -o SharedVector_class.o
/spack/apps/gcc/11.3.0/bin/gcc -I"/spack/2206/apps/linux-centos7-x86_64_v3/gcc-11.3.0/r-4.2.1-gof522x/rlib/R/include" -DNDEBUG -I’/home1/scastano/R/x86_64-pc-linux-gnu-library/4.2/S4Vectors/include’ -I’/home1/scastano/R/x86_64-pc-linux-gnu-library/4.2/IRanges/include’ -I/usr/local/include -fpic -g -O2 -c XDouble_class.c -o XDouble_class.o
/spack/apps/gcc/11.3.0/bin/gcc -I"/spack/2206/apps/linux-centos7-x86_64_v3/gcc-11.3.0/r-4.2.1-gof522x/rlib/R/include" -DNDEBUG -I’/home1/scastano/R/x86_64-pc-linux-gnu-library/4.2/S4Vectors/include’ -I’/home1/scastano/R/x86_64-pc-linux-gnu-library/4.2/IRanges/include’ -I/usr/local/include -fpic -g -O2 -c XInteger_class.c -o XInteger_class.o
/spack/apps/gcc/11.3.0/bin/gcc -I"/spack/2206/apps/linux-centos7-x86_64_v3/gcc-11.3.0/r-4.2.1-gof522x/rlib/R/include" -DNDEBUG -I’/home1/scastano/R/x86_64-pc-linux-gnu-library/4.2/S4Vectors/include’ -I’/home1/scastano/R/x86_64-pc-linux-gnu-library/4.2/IRanges/include’ -I/usr/local/include -fpic -g -O2 -c XRawList_comparison.c -o XRawList_comparison.o
/spack/apps/gcc/11.3.0/bin/gcc -I"/spack/2206/apps/linux-centos7-x86_64_v3/gcc-11.3.0/r-4.2.1-gof522x/rlib/R/include" -DNDEBUG -I’/home1/scastano/R/x86_64-pc-linux-gnu-library/4.2/S4Vectors/include’ -I’/home1/scastano/R/x86_64-pc-linux-gnu-library/4.2/IRanges/include’ -I/usr/local/include -fpic -g -O2 -c XRaw_class.c -o XRaw_class.o
/spack/apps/gcc/11.3.0/bin/gcc -I"/spack/2206/apps/linux-centos7-x86_64_v3/gcc-11.3.0/r-4.2.1-gof522x/rlib/R/include" -DNDEBUG -I’/home1/scastano/R/x86_64-pc-linux-gnu-library/4.2/S4Vectors/include’ -I’/home1/scastano/R/x86_64-pc-linux-gnu-library/4.2/IRanges/include’ -I/usr/local/include -fpic -g -O2 -c XVectorList_class.c -o XVectorList_class.o
/spack/apps/gcc/11.3.0/bin/gcc -I"/spack/2206/apps/linux-centos7-x86_64_v3/gcc-11.3.0/r-4.2.1-gof522x/rlib/R/include" -DNDEBUG -I’/home1/scastano/R/x86_64-pc-linux-gnu-library/4.2/S4Vectors/include’ -I’/home1/scastano/R/x86_64-pc-linux-gnu-library/4.2/IRanges/include’ -I/usr/local/include -fpic -g -O2 -c XVector_class.c -o XVector_class.o
/spack/apps/gcc/11.3.0/bin/gcc -I"/spack/2206/apps/linux-centos7-x86_64_v3/gcc-11.3.0/r-4.2.1-gof522x/rlib/R/include" -DNDEBUG -I’/home1/scastano/R/x86_64-pc-linux-gnu-library/4.2/S4Vectors/include’ -I’/home1/scastano/R/x86_64-pc-linux-gnu-library/4.2/IRanges/include’ -I/usr/local/include -fpic -g -O2 -c io_utils.c -o io_utils.o
io_utils.c:16:10: fatal error: zlib.h: No such file or directory
16 | #include <zlib.h>
| ^~~~~~~~
compilation terminated.
make: *** [/spack/2206/apps/linux-centos7-x86_64_v3/gcc-11.3.0/r-4.2.1-gof522x/rlib/R/etc/Makeconf:168: io_utils.o] Error 1
ERROR: compilation failed for package ‘XVector’
- removing ‘/home1/scastano/R/x86_64-pc-linux-gnu-library/4.2/XVector’
- installing source package ‘BiocParallel’ …
** using staged installation
checking for gcc… gcc
checking whether the C compiler works… no
configure: error: in /tmp/RtmpUKr6Hb/R.INSTALL72e8981365/BiocParallel': configure: error: C compiler cannot create executables See config.log’ for more details
ERROR: configuration failed for package ‘BiocParallel’
- removing ‘/home1/scastano/R/x86_64-pc-linux-gnu-library/4.2/BiocParallel’
- installing source package ‘RcppArmadillo’ …
** package ‘RcppArmadillo’ successfully unpacked and MD5 sums checked
** using staged installation
checking whether the C++ compiler works… no
configure: error: in /tmp/RtmpdnwhRc/R.INSTALL7e740cbd779/RcppArmadillo': configure: error: C++ compiler cannot create executables See config.log’ for more details
ERROR: configuration failed for package ‘RcppArmadillo’
- removing ‘/home1/scastano/R/x86_64-pc-linux-gnu-library/4.2/RcppArmadillo’
ERROR: dependency ‘openssl’ is not available for package ‘httr’
- removing ‘/home1/scastano/R/x86_64-pc-linux-gnu-library/4.2/httr’
ERROR: dependency ‘RCurl’ is not available for package ‘GenomeInfoDb’
- removing ‘/home1/scastano/R/x86_64-pc-linux-gnu-library/4.2/GenomeInfoDb’
ERROR: dependencies ‘XVector’, ‘GenomeInfoDb’ are not available for package ‘Biostrings’
- removing ‘/home1/scastano/R/x86_64-pc-linux-gnu-library/4.2/Biostrings’
ERROR: dependencies ‘GenomeInfoDb’, ‘XVector’ are not available for package ‘GenomicRanges’
- removing ‘/home1/scastano/R/x86_64-pc-linux-gnu-library/4.2/GenomicRanges’
ERROR: dependencies ‘httr’, ‘png’, ‘Biostrings’ are not available for package ‘KEGGREST’
- removing ‘/home1/scastano/R/x86_64-pc-linux-gnu-library/4.2/KEGGREST’
ERROR: dependencies ‘GenomicRanges’, ‘GenomeInfoDb’ are not available for package ‘SummarizedExperiment’
- removing ‘/home1/scastano/R/x86_64-pc-linux-gnu-library/4.2/SummarizedExperiment’
ERROR: dependency ‘KEGGREST’ is not available for package ‘AnnotationDbi’
- removing ‘/home1/scastano/R/x86_64-pc-linux-gnu-library/4.2/AnnotationDbi’
ERROR: dependencies ‘AnnotationDbi’, ‘XML’, ‘httr’ are not available for package ‘annotate’
- removing ‘/home1/scastano/R/x86_64-pc-linux-gnu-library/4.2/annotate’
ERROR: dependencies ‘AnnotationDbi’, ‘annotate’ are not available for package ‘genefilter’
- removing ‘/home1/scastano/R/x86_64-pc-linux-gnu-library/4.2/genefilter’
ERROR: dependencies ‘annotate’, ‘AnnotationDbi’ are not available for package ‘geneplotter’
- removing ‘/home1/scastano/R/x86_64-pc-linux-gnu-library/4.2/geneplotter’
ERROR: dependencies ‘GenomicRanges’, ‘SummarizedExperiment’, ‘BiocParallel’, ‘genefilter’, ‘geneplotter’, ‘RcppArmadillo’ are not available for package ‘DESeq2’
- removing ‘/home1/scastano/R/x86_64-pc-linux-gnu-library/4.2/DESeq2’
The downloaded source packages are in
‘/tmp/RtmpW5z5nJ/downloaded_packages’
Installation paths not writeable, unable to update packages
path: /spack/2206/apps/linux-centos7-x86_64_v3/gcc-11.3.0/r-4.2.1-gof522x/rlib/R/library
packages:
bslib, htmltools, MASS, pillar, rlang, sass, stringi
There were 18 warnings (use warnings() to see them)
install.packages(“openssl”)
Installing package into ‘/home1/scastano/R/x86_64-pc-linux-gnu-library/4.2’
(as ‘lib’ is unspecified)
trying URL ‘http://cran.r-project.org/src/contrib/openssl_2.0.2.tar.gz’
Content type ‘application/x-gzip’ length 1203211 bytes (1.1 MB)
==================================================
downloaded 1.1 MB
- installing source package ‘openssl’ …
** package ‘openssl’ successfully unpacked and MD5 sums checked
** using staged installation
Using PKG_CFLAGS=
--------------------------- [ANTICONF] --------------------------------
Configuration failed because openssl was not found. Try installing:
- deb: libssl-dev (Debian, Ubuntu, etc)
- rpm: openssl-devel (Fedora, CentOS, RHEL)
- csw: libssl_dev (Solaris)
- brew: openssl (Mac OSX)
If openssl is already installed, check that ‘pkg-config’ is in your
PATH and PKG_CONFIG_PATH contains a openssl.pc file. If pkg-config
is unavailable you can set INCLUDE_DIR and LIB_DIR manually via:
R CMD INSTALL --configure-vars=‘INCLUDE_DIR=… LIB_DIR=…’
-------------------------- [ERROR MESSAGE] ---------------------------
tools/version.c:1:10: fatal error: openssl/opensslv.h: No such file or directory
1 | #include <openssl/opensslv.h>
| ^~~~~~~~~~~~~~~~~~~~
compilation terminated.
ERROR: configuration failed for package ‘openssl’
- removing ‘/home1/scastano/R/x86_64-pc-linux-gnu-library/4.2/openssl’
Warning in install.packages :
installation of package ‘openssl’ had non-zero exit status
The downloaded source packages are in
‘/tmp/RtmpW5z5nJ/downloaded_packages’
library(pkgconfig)
install.packages(“openssl”)
Installing package into ‘/home1/scastano/R/x86_64-pc-linux-gnu-library/4.2’
(as ‘lib’ is unspecified)
trying URL ‘http://cran.r-project.org/src/contrib/openssl_2.0.2.tar.gz’
Content type ‘application/x-gzip’ length 1203211 bytes (1.1 MB)
==================================================
downloaded 1.1 MB
- installing source package ‘openssl’ …
** package ‘openssl’ successfully unpacked and MD5 sums checked
** using staged installation
Using PKG_CFLAGS=
--------------------------- [ANTICONF] --------------------------------
Configuration failed because openssl was not found. Try installing:
- deb: libssl-dev (Debian, Ubuntu, etc)
- rpm: openssl-devel (Fedora, CentOS, RHEL)
- csw: libssl_dev (Solaris)
- brew: openssl (Mac OSX)
If openssl is already installed, check that ‘pkg-config’ is in your
PATH and PKG_CONFIG_PATH contains a openssl.pc file. If pkg-config
is unavailable you can set INCLUDE_DIR and LIB_DIR manually via:
R CMD INSTALL --configure-vars=‘INCLUDE_DIR=… LIB_DIR=…’
-------------------------- [ERROR MESSAGE] ---------------------------
tools/version.c:1:10: fatal error: openssl/opensslv.h: No such file or directory
1 | #include <openssl/opensslv.h>
| ^~~~~~~~~~~~~~~~~~~~
compilation terminated.
ERROR: configuration failed for package ‘openssl’
- removing ‘/home1/scastano/R/x86_64-pc-linux-gnu-library/4.2/openssl’
Warning in install.packages :
installation of package ‘openssl’ had non-zero exit status
The downloaded source packages are in
‘/tmp/RtmpW5z5nJ/downloaded_packages’
Any help is appreciated,
Sofi
Sorry, I was able to download BiocManager but now I cannot install the DESeq2 package and the error message is what I posted above.
I made some changes to the RStudio app related to package installs, so you should be able to install most of these packages within RStudio now. Some packages may need special configuration to install.