I’m trying to set up a local library for perl modules but I’m getting a permission denied error - is there some other way I should be installing perl modules?
@ckenkel There’s more than one way to do this, but I’ve installed the local::lib module for Perl which will be useful in any case. After entering module load perl, you can then use the cpanm command, for example, to install into a specified directory. Something like:
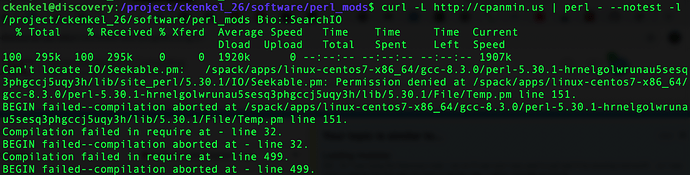
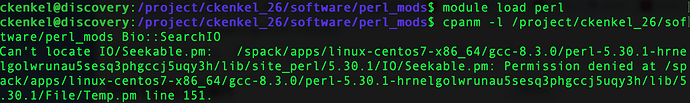
cpanm -l /project/ckenkel_26/software/perl_mods Bio::SearchIO
You would also need to add a line at the top of your Perl scripts like:
use local::lib '/project/ckenkel_26/software/perl_mods';
so that Perl can find the library.
Still getting the same error with the seekable module and trying to install that one locally doesn’t work either (same error)
Ah, that module directory didn’t have r-x permissions for general users. I’ve changed it now, so please try again later today and let me know if it works.
seems like this might be a problem with other modules - the new error is:
Can’t locate ExtUtils/Manifest.pm in @INC
and same thing, trying to do a local install of ExtUtils::MakeMaker requires this module
Hmm, so it seems that on CentOS a lot of standard modules are not installed by default, in contrast to other Linux distributions. Sorry about that. I’ll look into this more and install those modules. I’ve also installed perlbrew in case you want to install your own separate version of Perl to use.
@ckenkel I’m actually not sure why you were getting the error message Can’t locate ExtUtils/Manifest.pm in @INC as it did exist. Could you share the output after entering these commands?
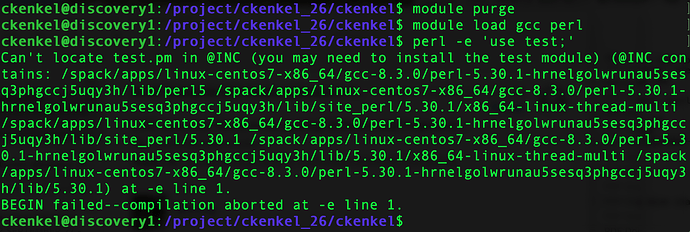
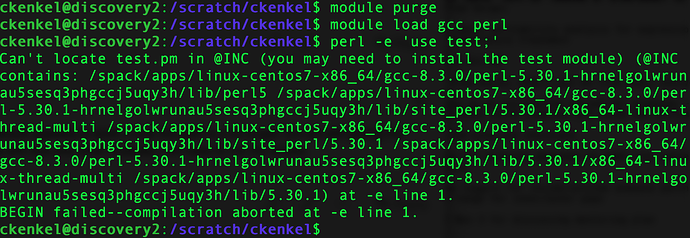
module purge
module load gcc perl
perl -e 'use test;'
It should be a similar error about can’t locate in @INC , but I’m wondering what directories it’s looking in for you.
I’ve just installed BioPerl as its own module and it will be available on Discovery later today, so you can use that instead of a local::lib. You can load it with:
module purge
module load gcc perl perl-bioperl
And then Bio::SearchIO and the other BioPerl modules will be available to use. Let me know if that works for you.
Thanks for pursuing this - and good timing - was going to work on a local perl install this am - would be glad to spend my time on something else insead! The error message is below.
I have my module commands written into my .bashrc - it would be nice not to have to purge them every time to get perl to run, but understand the need for troubleshooting. Am happy to share that list as well if you would like to see it.
Thanks for the bioperl install as well, but I will likely need other modules as well - it would be great to get this local install issue sorted if possible!
Also - the bioperl module appears to work in that the path is a lot longer, but am still getting the same module errors with this and other modules I will need to run my scripts.
The module purge was just for testing. Those are the correct directories where Perl should look. Do you get the same error message, with the same directories listed, without the module purge (and a fresh login)? And the same error message when loading a module that is installed? For example, perl -e 'use ExtUtils::MakeMaker;' should not produce an error.
Yes, please share elements of your .bashrc that might affect this, such as modules loaded or environment variables set.
I may have found the issue. The first directory listed in @INC actually did not exist (/spack/apps/linux-centos7-x86_64/gcc-8.3.0/perl-5.30.1-hrnelgolwrunau5sesq3phgccj5uqy3h/lib/perl5), so perhaps Perl simply stops looking after it can’t find that directory. This wasn’t an issue for me though, so I’m not sure. I’ve created that directory anyways, even though it is not used, and synced it over to Discovery. Try again later today and see if that changes anything.
Nope. Same error for both a fresh login and after the module purge and reload for ‘test’ (at least superficially I think so, have not gone line by line to compare)
I don’t get any error for the ExtUtils::MakeMaker for either the fresh login or module purge and reload of only gcc and perl
There doesn’t appear to be a way to attach a file but you should be able to view my bashrc edits using this link https://drive.google.com/file/d/1pjw7HlhLjOmF7XdTdzXzu56JGV7mxobn/view?usp=sharing
That’s fine, perl -e 'use test;' should produce an error as there is no test module. I just used that as an error test. If you get no error for ExtUtils::MakeMaker, then it sounds like the @INC issue is fixed. Are you able to install a module locally now without the @INC error?
You will likely get an error trying to install Bio::SearchIO, but that’s a separate issue related to one of the XML dependency modules. Try installing Statistics::Descriptive, for example.
okay - worked for Statistics::Descriptive! thanks!